As
mentioned earlier one of interesting features of metal nanoparticles is their
fascinating optical properties “Hyperlink Missing”. So to control changes in a
solution during the production of nanoparticles is to monitor changes in the
solution optical properties.
Nanoparticles
for the detection of specific DNA
The
nanoparticles shown in following figure are designed to detect the presence of
specific DNA strand. For this purpose the nanoparticles are modified via a
thiol bond with single stranded DNA complementary to the target DNA. The
modification of the nanoparticles affects the amount of light absorbed by the
solution, compared to the un-modified nanoparticles, which have a
characteristics peak at 560 nm.
When
target DNA analyte is added to the solution, aggregation occurs. Nanoparticles
move closer together and form clusters. The formation of nanoparticle clusters
results in changes of absorbance of the solution. The location of the
absorbance peak is an indication of average nanoparticle size. When peak is
shifted to higher wavelength in this case from 560nm to 600 nm it means that
the average size of the particles in the solution is larger due to the
aggregation. The width of the peak much
broader after aggregation. This means a higher distribution of nanoparticle
size compared to the bare nanoparticles. The presence of a target analyte can
be therefore determined by monitoring the absorbance of the solution.
Monitoring
the change in optical properties at specific wavelengths, it is possible to
examine the sensing properties of these sensors by applying following
procedure:
The signal obtained at 560 nm relates to the DNA molecules
and the signal obtained at 700 nm relates to aggregates of DNA with gold nanoparticles.
When the solution is heated to 80°C the target DNS
dissolves in solution and the relevant signal at 560 nm increases. This results
in decrease in the aggregate peak at 700 nm, as the DNA/Au aggregates
dissociate to individual nanoparticles. This process is repeatable when heating
and cooling the system. The heating and cooling is also visible by a naked eye
by looking at solution color changes.
In
general the DNA-capped gold nanoparticles are promising approach for the
detection of DNA strands by controlling solution temperature of DNA-modified
nanoparticles.
Various
examples of nanoparticles used for DNA detection
When
sensor meets its target DNA, the color of the drop changes due to aggregation.
This system does not require sophisticated equipment for analysis and is
appropriate mainly to determine the presence of analyte, not its concentration.
Detecting small concentrations of target DNA is more challenging.
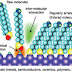
In
figure X-B the gold nanoparticles modified with short, single-stranded DNA to
indicate the presence of particular DNA sequence that is hybridized on a
transparent substrate in a three-component sandwich assay format. At high
target DNA concentrations, many nanoparticles are attached to the transparent
surface. The high concentration of nanoparticles causes a dramatic change in
the color and the substrate looks pink. At lower target concentrations, the
attached nanoparticles cannot be visualized with the naked eye. To facilitate
visualization of nanoparticles labels that are hybridized to the array surface,
planting silver ions on the gold nanoparticles is used for signal
amplification. Using silver ions very low coverage of nanoparticles can be
visualized by a simple scanner or by using naked eye.
In
figure X-C the attachment of nanoparticles to a substrate in a three-component
sandwich assay format is detected by the change in scattered light. A more applicable approach is monitoring the
electrical changes of a sensor while the sensing mechanism is similar to the
previous examples as seen in figure X-D. The system consist of isolating
substrate with two electrodes for measuring the electrical resistance. On the
top of isolating substrate the nanoparticles are assembled in a low
concentration so there is no direct connection between the nanoparticles and
therefore the electrical resistance is very high. If DNA is captured, silver
will plate the nanoparticles. This plating will contact between the
nanoparticles and the electrical resistance will dramatically decrease. By
measuring the changes in the electrical resistance we can obtain and monitor
sensing signal.
There
are different approaches that utilize similar sensing mechanisms, but use different
characterization methods that have been developed and tested to detect the
attachment of nanoparticles to a substrate.
Monitor
the surface enhanced Raman spectroscopy (SERS) signal as seen in figure X-E
Monitor
the diffraction of laser beam, when the laser beam is directed towards the
nanoparticles that are attached to the
surface as seen in figure X-F
Magnetic
nanoparticles
To
detect tiny amounts of targeted molecules inside the human body or in
complicated environments relies on magnetic nanoparticles. Magnetic micro
particle probes with antibodies that specifically bind a target of interest and
nanoparticle probes that are encoded with DNA that is unique to the protein
target of interest and antibodies that can sandwich the target captured by the
micro particle probes.
Magnetic
separation of the complex probes and target is followed by de-hybridization of
the DNA on the nanoparticle probe surface, thus allowing one to determine the
presence of the target protein by identifying the DNA sequence released from
the nanoparticle probe.








0 comments:
Post a Comment